|
Stock number (links to data sheet)
|
Strain name
(links to expression data)
|
Promoter (species)
|
Expected site of expression
|
Expression data thumbnail
|
|
006149
|
B6.Cg-Tg(ACTA1-cre)79Jme/J
|
ACTA1 (human)
|
Cre activity is restricted to adult striated muscle fibers and embryonic striated muscle cells of the somites and heart.
|
  |
|
010803
|
B6;FVB-Tg(Adipoq-cre)1Evdr/J
|
Adipoq, adiponectin, C1Q and collagen domain containing (mouse)
|
Cre recombinase activity is expected in white adipose tissue (WAT) and brown adipose tissue (BAT)
|
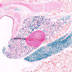 |
|
012899
|
STOCK Agrptm1(cre)Lowl/J
|
Agrp (mouse)
|
ArGP neurons in the hypothalamus
|
 |
|
003574
|
B6.Cg-Tg(Alb-cre)21Mgn/J
|
Alb, albumin (rat)
|
Cre recombinase expression is expected in the Liver
|
 |
|
016832
|
B6.FVB(129)-Tg(Alb1-cre)1Dlr/J
|
Alb1, albumin (mouse)
|
Cre recombinase expression is expected in the Liver
|
 |
|
007915
|
129S.FVB-Tg(Amh-cre)8815Reb/J
|
Amh (mouse)
|
Testis Sertoli cell-specific promoter elements of the anti-Mullerian hormone (Amh) gene
|
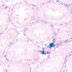 |
|
006881
|
B6.Cg-Tg(Aqp2-cre)1Dek/J
|
Aqp2 (mouse)
|
Transgenic cre activity, directed by the mouse aquaporin 2 promoter, is observed in kidney cells (collecting duct, left) and testes (sperm, right).
|
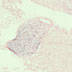 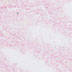 |
|
010774
|
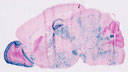
B6(Cg)-Calb2tm1(cre)Zjh/J
|
Calb2, calbindin 2
|
Cre activity is expected in calretinin interneurons in the brain and cortex driven by the endogenous Calb2 promoter/enhancer elements
|
 |
|
005359
|
B6.Cg-Tg(Camk2a-cre)T29-1Stl/J
|
Camk2a, calcium/calmodulin-dependent protein kinase II alpha (mouse)
|
Cre recombinase expression is expected in the forebrain, specifically the CA1 pyramidal cell layer in the hippocampus
|
 |
|
012706
|
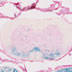
STOCK Ccktm1.1(cre)Zjh/J
|
Cck, cholecystokinin (mouse)
|
Cre recombinase activity is expected in cholecystokinin positive neurons (interneurons) of the cortex.
Expression also seen in Adult spinal cord and embryonic day 15.5 spinal cord and heart (bottom left and right).
|
   |
|
008520
|
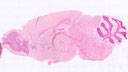
B6.Cg-Tg(CD2-cre)4Kio/J
|
CD2, CD2 molecule (human)
|
Cre recombinase activity is expected in T cells and B cells (all committed B cell and T cell progenitors)
|
 |
|
004126
|
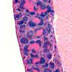
C.Cg-Cd19tm1(cre)Cgn/Ighb/J
|
Cd19
|
Cre recombinase expression is expected in B cells
|
 |
|
006137
|
B6.Cg-Tg(Cdh5-cre)7Mlia/J
|
Cdh5, cadherin 5
|
Embryonic and adult cre recombinase activity is reported in endothelium of developing and quiescent vessels of all organs examined, as well as within a subset of hematopoietic cells
|
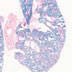 |
|
012237
|
B6.Cg-Tg(Cdh16-cre)91Igr/J
|
Chd16 (mouse)
|
In the adult mouse expression is limited to the renal tubules especially the collecting ducts, loops of Henle and distal tubules.
|
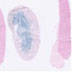 |
|
006410
|
B6;129S6-Chattm2(cre)Lowl/J
|
Chat, choline acetyltransferase (mouse)
|
Cre recombinase activity is reported in all cholinergic neurons
|
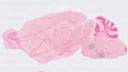 |
|
006475
|
B6.FVB(129S4)-Tg(Ckmm-cre)5Khn/J
|
Ckmm (mouse)
|
These transgenic mice have the Cre recombinase gene driven by the muscle creatine kinase promoter, and Cre activity is observed in skeletal and cardiac muscle.
|
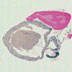 |
|
010910
|
STOCK Corttm1(cre)Zjh/J
|
Cort, cortistatin
|
Cre recombinase expression is expected in Cort-expressing cells (CST positive neurons)
|
 |
|
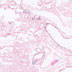
012704
|
B6(Cg)-Crhtm1(cre)Zjh/J
|
Crh, corticotropin releasing hormone
|
Cre recombinase activity is expected in CRH positive neurons
|
 |
|
JR: 8538
|
B6.Cg-Tg(Cspg4-cre/Esr1*)BAkik/J
|
Cspg4
(Mouse)
|
DI reports NG2-expressing glia (polydendrocytes, oligodendrocyte progenitor cells) in the central nervous system and NG2-expressing cells in other organs. Donating Investigator reports staining in Corpus Callosum with GFP antibody.
In our hands Lac Z staining was only observed at low level in the CNS and higher levels in other tissues such as testes and blood vessels (see MGI link).
|


|
|
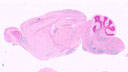
008839
|
B6;C3-Tg(Cyp39a1-cre)1Aibs/J
|
Cyp39a1, cytochrome P450, family 39, subfamily a, polypeptide 1 (mouse)
|
Cre recombinase expression is directed to cerebral cortex, hippocampus, striatum, olfactory bulb, and cerebellum
|
 |
|
008199
|
STOCK Tg(dlx6a-cre)1Mekk/J
|
dlx6a, distal-less homeobox gene 6a
|
Cre recombinase activity is expected in GABAergic forebrain neurons
|
 |
|
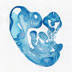
003314
|
FVB/N-Tg(EIIa-cre)C5379Lmgd/J
|
Ella, adenovirus (adenovirus)
|
Cre recombinase activity is expected in a wide range of tissues, including the germ cells that transmit the genetic alteration to progeny
|
 |
|
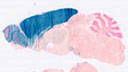
005628
|
B6.129S2-Emx1tm1(cre)Krj/J
|
Emx1, empty spiracles homolog 1 (Drosophila)
|
Cre recombinase activity is reported in neurons of the neocortex and hippocampus, and in the glial cells of the pallium, mimicking the pattern of expression of the endogenous gene
|
 |
|
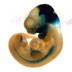
007916
|
STOCK En1tm2(cre)Wrst/J
|
En1, engrailed 1
|
Cre recombinase activity is expected in spinal cord V1 interneurons, the embryonic mesencephalon and rhombomere 1 by E9, as well as in the ventral ectoderm of the limbs, in a subset of somite cells, and some mesoderm-derived tissues.
|
 |
|
005069
|
B6.Cg-Tg(Fabp4-cre)1Rev/J
|
Fabp4, fatty acid binding protein 4
|
Cre recombinase activity is reported in brown and white adipose tissue.
|
  |
|
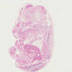
012463
|
B6;129S4-Foxd1tm1(GFP/cre)Amc/J
|
Foxd1 (mouse)
|
When Foxd1 expression is activated, expression is observed during kidney development in metanephric mesenchyme in cells fated to become stromal cells of the kidney. When FoxD1GC mice are bred with mice containing loxP-flanked sequence, Cre-mediated recombination will result in deletion of the floxed sequences in the Foxd1-expressing cells of the offspring. Expression also seen in multiple organs throughout the body.
|
 |
|
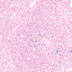
008694
|
NOD/ShiLt-Tg(Foxp3-EGFP/cre)1cJbs/J
|
Foxp3 (mouse)
|
Expression is specific toCd4+Cd25<high>Cd127<low>T cells from the lymph nodes, spleen and thymus. Expression also seen in ovary.
|
 |
|
010802
|
STOCK Gad2tm2(cre)Zjh/J
|
Gad2, glutamic acid decarboxylase 2
|
Cre recombinase activity is expected in Gad2 positive neurons
|
 |
|
004600
|
FVB-Tg(GFAP-cre)25Mes/J
|
GFAP, glial fibrillary acidic protein (human)
|
Cre recombinase activity is reported in the central nervous system, affecting astrocytes, oligodendroglia, ependyma and some neurons; also periportal cells of the liver
|
 |
|
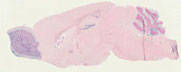
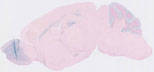
012886
|
B6.Cg-Tg(Gfap-cre)73.12Mcs/J
|
Gfap (mouse)
|
GFAP-Cre transgenic mice from founder line 73.12 have Cre recombinase expression directed by the mouse glial fibrillary acidic protein promoter. Cre expression is observed in astrocytes in the brain and spinal cord, as well as postnatal and adult GFAP-expressing neural stem cells and their progeny in the brain. Other expression observed in cartilage primordium at e15.5; and in thymus, myocardium, eye lens, peripheral nerves embedded in bladder and intestinal muscle of adults.
|
 |
|
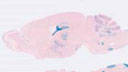
012887
|
B6.Cg-Tg(Gfap-cre)77.6Mvs/J
|
Gfap (mouse)
|
Cre recombinase activity is targeted to most astrocytes throughout the healthy brain and spinal cord and to essentially all astrocytes after Central Nervous System (CNS) injury. Cre recombinase activity is also targeted to a subpopulation of the adult stems in the subventricular zone.
|
 |
|
006474
|
C57BL/6-Tg(Grik4-cre)G32-4Stl/J
|
Grik4, glutamate receptor, ionotropic, kainate 4 (mouse)
|
Cre recombinase activity is reported at 14 days old in area CA3 of the hippocampus, and at 8 weeks of age, recombination is observed in nearly 100% of pyramidal cells in area CA3; recombination is also observed in other brain areas, but at distinctly lower frequencies
|
 |
|
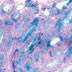
008870
|
C57BL/6-Tg(Hspa2-cre)1Eddy/J
|
Hspa2, heat shock protein 2 (mouse)
|
Cre recombination is expected to occur in leptotene/zygotene spermatocytes
|
 |
|
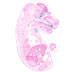
003573
|
B6.Cg-Tg(Ins2-cre)25Mgn/J
|
Ins2, insulin 2 (rat)
|
Cre recombinase activity is expected in pancreatic beta cells, as well as the hypothalamus
|
 |
|
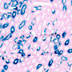
008068
|
B6.Cg-Tg(Itgax-cre)1-1Reiz/J
|
Itgax, integrin alpha X (mouse)
|
Cre recombinase activity is expected in CD8-, CD8+dendritic cells, tissue derived dendritic cells from lymph nodes, lung and epidermis and plasmacytoid dendritic cells
|
 |
|
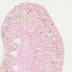
008781
|
B6.Cg-Tg(Kap-cre)29066/2Sig/J
|
Kap (mouse)
|
Cre recombinase expression is detected in the proximal tubule cells of the renal cortex in male mice. Female mice do not express the transgene unless treated with androgen (testosterone).Note: images shown are from female mice. Expression also observed in uterus and liver.
|
 |
|
004782
|
STOCK Tg(KRT14-cre)1Amc/J
|
KRT14, keratin 14 (human)
|
Cre recombinase activity is reported in skin, the oral ectoderm including the dental lamina at 11.75 d.p.c., and the dental epithelium by 14.5 d.p.c.
|
 |
|
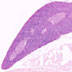
003802
|
B6.Cg-Tg(Lck-cre)548Jxm/J
|
Lck, lymphocyte protein tyrosine kinase (mouse)
|
Cre recombinase activity is reported in thymocytes
|
 |
|
012837
|
B6.Cg-Tg(Lck-cre)3779Nik/J
|
Lck (mouse)
|
These mice are useful for studying the consequences of mutations induced after positive selection in the thymus.
|
 |
|
008320
|
B6.129-Leprtm2(cre)Rck/J
|
Lepr (mouse)
|
Cre activity is demonstrable in the hypothalamus (arcuate, dorsomedial, lateral, and ventromedial nuclei), limbic and cortical brain regions (basolateral amygdaloid nucleus, piriform cortex, and lateral entorhinal cortex), and retrosplenial cortex.
|
 |
|
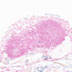
012601
|
B6;129P2-Lyve1tm1.1(EGFP/cre)Cys/J
|
Lyve1 (mouse)
|
This strain is useful for conditional ablation of genes in the lymphatic endothelium and studies of lymphatic development, function and lymphangiogenesis.
|
  |
|
004781
|
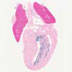
B6.129P2-Lyz2tm1(cre)Ifo/J
|
Lyz2, Lysozyme 2 (mouse)
|
Cre recombinase activity is reported in myeloid cells, including monocytes, mature macrophages and granulocytes
|
 |
|
003553
|
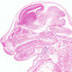
STOCK Tg(MMTV-cre)4Mam
|
MMTV |
High levels of recombination have been detected in the virgin and lactating mammary gland, salivary gland, seminal vesicle, skin, erythrocytes, B cells and T cells. Little background recombination was observed in the lung, kidney, liver and brain tissues (less than 10%).
|
   |
|
006600
|
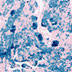
B6.129S1-Mnx1tm4(cre)Tmj/J
|
Mnx1, motor neuron and pancreas homeobox 1 (mouse)
|
Cre recombinase activity is reported in motor neurons
|
 |
|
007893
|
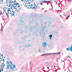
B6.129S4-Myf5tm3(cre)Sor/J
|
Myf5, myogenic factor 5
|
Cre recombinase activity is expected in skeletal muscle and the dermis, and in several ectopic locations
|
 |
|
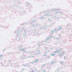
011038
|
B6.FVB-Tg(Myh6-cre)2182Mds/J
|
Myh6 (mouse)
|
Cardiac tissue
|
 |
|
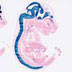
003771
|
B6.Cg-Tg(Nes-cre)1Kln/J
|
Nes, nestin (rat)
|
Cre recombinase activity is reported in the central and peripheral nervous system and a few isolated kidney and heart cells
|
 |
|
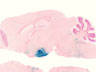
006333
|
B6.FVB(Cg)-Tg(Neurog3-cre)C1Able/J
|
Neurog3, neurogenin 3, (rat)
|
Cre recombinase activity is expected in islets of the adult pancreas, small intestine enteroendocrine cells, endocrine portions of the stomach, all pancreatic endocrine cells, and in some non-endocrine intestinal cells
|
 |
|
008661
|
C57BL/6J-Tg(Nkx2-1-cre)2Sand/J
|
Nkx2-1
|
Cre recombinase activity is directed to brain interneuron progenitors, developing lung, thyroid, and pituitary by the Nkx2.1 promoter/enhancer regions
|
 |
|
008205
|
B6.Cg-Tg(NPHS2-cre)295Lbh/J
|
NPHS2 (human)
|
Cre-recombinase activity is reported in podocytes during late capillary loop stage of glomerular development and persists in podocytes of mature glomeruli
|
 |
|
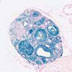
006364
|
FVB-Tg(Nr5a1-cre)2Lowl/J
|
Nr5a1, Nuclear receptor subfamily 5 group A member 1 (mouse)
|
Cre recombinase activity is reported in Ventromedial Hypothalamus, Cortex, Adrenal Gland, Pituitary Gland and Gonads
|
 |
|
006668
|
B6;129P2-Omptm4(cre)Mom/MomJ
|
Omp, Olfactory Marker Protein (mouse)
|
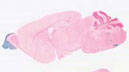
Cre recombinase activity is reported in mature olfactory sensory neurons
|
 |
|
005549
|
B6;129-Pax3tm1(cre)Joe/J
|
Pax3, paired box gene 3
|
Cre recombinase activity is expected in the dorsal neural tube and somites of E9 to 11.5 embryos and in the cardiac neural crest cells and colonic epithelia of E11.5 embryos
|
 |
|
008535
|
C57BL/6-Tg(Pf4-cre)Q3Rsko/J
|
Pf4, platelet factor 4 (mouse)
|
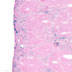
Cre activity is expected in the majority of megakaryocytes
|
 |
|
005965
|
STOCK Tg(Pomc1-cre)16Lowl/J
|
Pomc1 (mouse)
|
Cre recombinase activity is reported in POMC neurons in the arcuate nucleus of the hypothalamus and scattered in the dentate gyrus of the hippocampus
|
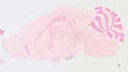 |
|
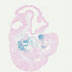
008827
|
B6.Cg-Tg(Prdm1-cre)1Masu/J
|
Prdm1 (mouse)
|
Cre-mediated recombination is detected in 55-76%
of primordial germ cells.
|
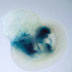  |
|
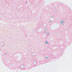
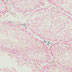
003328
|
129S/Sv-Tg(Prm-cre)58Og/J
|
Prm (mouse)
|
A cre recombinase coding sequence, driven by the mouse protamine 1 promoter (Prm) in this transgenic strain, mediates the efficient recombination of a cre target transgene in the male germ line. Other low level expression seen in kidney glomeruli, brain and salivary gland
|
  |
|
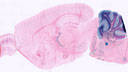
008069
|
B6;129P2-Pvalbtm1(cre)Arbr/J
|
Pvalb, parvalbumin
|
Cre activity is expected in neurons that express parvalbumin, such as interneurons in the brain and proprioceptive afferent sensory neurons in the dorsal root ganglia
|
 |
|
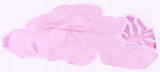
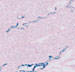
009613
|
B6;C3-Tg(Scnn1a-cre)3Aibs/J
|
Scnn1a (mouse)
|
Cre expression directed to cortex, thalamus, midbrain, and cerebellum.
|
  |
|
005622
|
B6.Cg-Shhtm1(EGFP/cre)Cjt/J
|
Shh, sonic hedgehog
|
Cre activity is expected to follow endogenous Shh expression patterns
|
 |
|
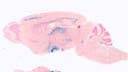
006395
|
STOCK Tg(Sim1-cre)1Lowl/J
|
Sim1, single-minded homolog 1 (Drosophila)(mouse)
|
Cre recombinase activity is expected in all areas that endogenously express Sim1, including paraventricular hypothalamus and other parts of the brain
|
 |
|
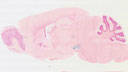
006660
|
B6.SJL-Slc6a3tm1.1(cre)Bkmn/J
|
Slc6a3, solute carrier family 6 (neurotransmitter transporter, dopamine), member 3
|
Cre recombinase activity is expected in dopaminergic cell groups (substantia nigra (SN) and ventral tegmental area (VTA), as well as in the retrorubral field)
|
 |
|
016963
|
STOCK Slc17a6tm2(cre)Lowl/J
|
Slc17a6 (mouse)
|
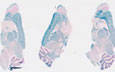
Cre recombinase expression directed to excitatory glutamatergic neuron cell bodies
|

|
|
013044
|
STOCK Ssttm2.1(cre)Zjh/J
|
Sst, somatostatin
|
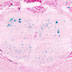
Cre recombinase activity is expected in somatostatin positive neurons (including dendritic inhibitory interneurons such as Martinotti cells and Oriens-Lacunosum-Moleculare cells)
|
 |
|
008208
|
STOCK Tg(Stra8-cre)1Reb/J
|
Stra8 (mouse)
|
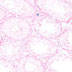
Stra8-cre transgenic mice may be useful in generating conditional knockouts in postnatal, premeiotic, male germ cells for studying spermatogenesis.
|
  |
|
003966
|
B6.Cg-Tg(Syn1-cre)671Jxm/J
|
Syn1 (rat)
|
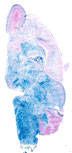
Cre recombinase activity is detected in neuronal cells, including brain, spinal cord and DRGs, as early as E12.5, as well as in neurons in adult.
|
 |
|
004746
|
STOCK Tg(Tagln-cre)1Her/J
|
Tagln, transgelin (mouse)
|
Cre recombinase activity is expected in smooth muscle
|
 |
|
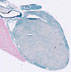
006878
|
B6.129S6-Taglntm2(cre)Yec/J
|
Tagln (mouse)
|
Cre recombinase activity is shown in adult smooth muscle cells (such as arteries, veins, and visceral organs) and cardiac myocytes, but activity is not observed in the same embryonic tissues.
|
 |
|
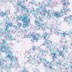
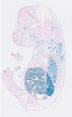
008863
|
B6.Cg-Tg(Tek-cre)1Ywa
|
Tek (mouse)
|
Uniform expression in endothelial cells during embryogenesis and adulthood.
|
  |
|
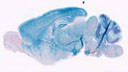
006143
|
FVB/N-Tg(Thy1-cre)1Vln/J
|
Thy1 (mouse)
|
Cre recombinase activity is expected in neurons of the cortex and hippocampus.
|
 |
|
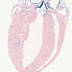
008712
|
B6.129X1-Twist2tm1.1(cre)Dor/J
|
Twist2, twist basic helix-loop-helix transcription factor 2
|
Cre recombinase activity is expected in mesoderm as early as embryonic day 9.5, in mesodermal tissues such as branchial arches and somites, and in condensed mesenchyme-derived chondrocytes and osteoblasts.
|
 |
|
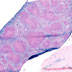
008610
|
B6.Cg-Tg(Vav1-cre)A2Kio/J
|
Vav1 (mouse)
|
Using crosses to a reporter strain, variegated germline (testis and ovaries), and heart and gut expression is also reported. When bred with mice containing a loxP-flanked sequence of interest, Cre-mediated recombination will result in deletion of the floxed sequence(s) in the offspring. These Vav1-Cre transgenic mice may be useful for generating conditional mutations in hematopoietic cells.
|
 |
|
004586
|
B6.SJL-Tg(Vil-cre)997Gum/J
|
Vil1, villin 1 (mouse)
|
Cre recombinase activity is expected in villi and crypts of the small and large intestine
|
 |
|
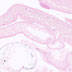
010908
|
STOCK Viptm1(cre)Zjh/J
|
Vip, vasoactive intestinal polypeptide
|
Cre recombinase activity has been reported in some GABAergic interneurons
|
 |
|
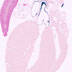
009107
|
B6.Cg-Tg(Wnt1-cre)11Rth Tg(Wnt1-GAL4)11Rth/J
|
Wnt1, wingless-related MMTV integration site 1 (mouse)
|
Cre recombinase activity is expected in embryonic neural tube, midbrain, dorsal and ventral midlines of the midbrain and caudal diencephalon, the mid-hindbrain junction and dorsal spinal cord
|
 |
|
022137
|
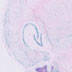
129S4.Cg-Tg(Wnt1-cre)2Sor/J
|
Wnt1 (mouse)
|
Applications in studies of cell lineage tracing in the developing neural crest and midbrain.
|
  |
Facebase Tools Cre Strains:
|
|
Stock number(links to data sheet)
|
Strain name
(links to expression data)
|
Promoter (species)
|
Expected site of expression
|
Expression data thumbnail
|
|
018151
|
C57BL/6N-Krt17tm1(cre,Cerulean)Murr/GrsrJ
|
Krt17, keratin 17 (mouse)
|
Cre recombinase expression is driven by the endogenous keratin 17 (Krt17) promoter/enhancer elements
|

|
|
009388
|
B6;129S1-Osr2tm2(cre)Jian/J
|
Osr2, odd-skipped related 2 (Drosophila), mouse, laboratory
|
Cre recombinase activity is reported in developing palate and urogenital tract
|

|
|
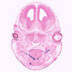
018791
|
C57BL/6J-Tg(Trp63,-cre,-Cerulean)3Grsr/GrsrJ
|
Trp63, transformation related protein 63 (mouse)
|
Cre recombinase expression is driven by Trp63promoter/enhancer elements
|
|
|
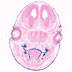
018792
|
C57BL/6J-Tg(Trp63,-cre,-Cerulean)4Grsr/GrsrJ
|
Trp63, transformation related protein 63 (mouse)
|
Cre recombinase expression is driven by Trp63promoter/enhancer elements
|
|
|
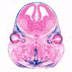
005584
|
B6.Cg-Tg(Prrx1-cre)1Cjt/J
|
Prrx1, paired related homeobox 1 (rat)
|
Cre recombinase activity is expected in early limb bud mesenchyme and in a subset of craniofacial mesenchyme, along with limited female germline expression
|
|
|
018754
|
C57BL/6J-Tg(Tbx22,-cre,-mCherry)1Grsr/GrsrJ
|
Tbx22, T-box transcription factor 22 (mouse)
|
Cre recombinase expression is driven by Tbx22promoter/enhancer elements
|

|
|
012719
|
STOCK Tgfb3tm1(cre)Vk/J
|
Tgfb3, transforming growth factor, beta 3 (mouse)
|
Cre recombinase activity is expected in the heart, pharyngeal arches, otic vesicle, mid brain, limb buds, midline palatal epithelium, and whisker follicles during embryo and fetus development
|

|
|
009107
|
B6.Cg-Tg(Wnt1-GAL4)11Rth Tg(Wnt1-Cre)11Rth
|
Wnt1, wingless-related MMTV integration site 1 (mouse)
|
Cre recombinase activity is expected in embryonic neural tube, midbrain, caudal diencephalon, the mid-hindbrain junction, dorsal spinal cord, and neural crest cells
|
 |
Inducible Strains:
|
|
Stock number(links to data sheet)
|
Strain name
(links to expression data)
|
Promoter (species)
|
Expected site of expression
|
Expression data thumbnail
|
|
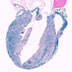
004682
|
B6.Cg-Tg(CAG-cre/Esr1)5Amc/J
|
ACTB, actin, beta (chicken)
|
Tamoxifen-inducible Cre recombinase expression is expected in most tissue types
|
|
|
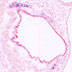
006774
|
FVB-Tg(Col2a1-cre/ERT)KA3Smac/J
|
Col2a1, collagen, type II, alpha 1 (mouse)
|
Tamoxifen-inducible cre expression is expected in cells of the chondrogenic lineage (cartilage) during embryogenesis and postnatally.
|
|
|
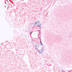
010705
|
B6(Cg)-Dlx5tm1(cre/ERT2)Zjh/J
|
Dlx5, distal-less homeobox 5
|
Tamoxifen-inducible Cre recombinase activity is expected in the cortex
|
|
|
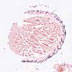
005107
|
STOCK Tg(KRT14-cre/ERT)20Efu/J
|
KRT14, keratin 14 (human)
|
Tamoxifen-inducible Cre recombinase expression is expected in keratinocytes
|
|
|
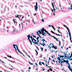
008875
|
B6.129P2-Lgr5tm1(cre/ERT2)Cle/J
|
Lgr5, leucine rich repeat containing G protein coupled receptor 5
|
Tamoxifen-inducible Cre recombinase expression is expected in crypt base columnar cells in small intestine (stem cells of the small intestine) and colon
|
|
|
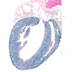
005657
|
B6.FVB(129)-Tg(Myh6-cre/Esr1)1Jmk/J
|
Myh6, myosin, heavy polypeptide 6,(mouse)
|
Tamoxifen-inducible Cre recombinase expression is expected in developing and adult heart
|
|
|
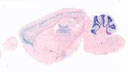
005975
|
B6.Cg-Tg(Plp1-cre/ERT)3Pop/J
|
Plp1, proteolipid protein (myelin) 1 (mouse)
|
Tamoxifen-inducible Cre recombinase expression is expected in oligodendrocytes and Schwann cells
|
|
|
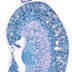
008085
|
B6.Cg-Ndor1Tg(UBC-cre/ERT2)1Ejb/2J
|
UBC, ubiquitin C (human)
|
Tamoxifen-inducible Cre recombinase expression is expected in all tissue types
|
|
|
009103
|
B6;C3-Tg(Wfs1-cre/ERT2)3Aibs/J
|
Wfs1, Wolfram syndrome 1 homolog (human)
|
Tamoxifen-inducible Cre recombinase expression is directed to cortex, hippocampus, striatum, thalamus and cerebellum only following tamoxifen administration
|
|