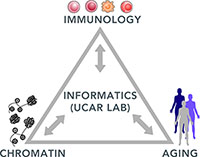
The Ucar Lab
Develops computational methods to study the epigenetic regulation of gene expression and its implications in important human conditions including aging and common diseases.
Principal Investigator
Location
Topics
Our Research Focus
Genomic signatures of immune system aging
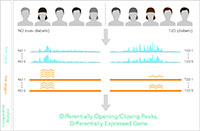
 The functional decline of the immune system with aging, i.e., immunosenescence, has been associated with increased disease susceptibility, infections, and poor response to treatments and vaccination in elderly. However, we have yet to discover which genes, enhancers and regulatory interactions in which immune cells can explain the functional decline of immune functions with aging and whether these changes are similar between sexes and different populations. In collaboration with
and UCHC Center on Aging we study chromatin signatures of human immune system aging and to quantify the clinical implications of these signatures.
The functional decline of the immune system with aging, i.e., immunosenescence, has been associated with increased disease susceptibility, infections, and poor response to treatments and vaccination in elderly. However, we have yet to discover which genes, enhancers and regulatory interactions in which immune cells can explain the functional decline of immune functions with aging and whether these changes are similar between sexes and different populations. In collaboration with
and UCHC Center on Aging we study chromatin signatures of human immune system aging and to quantify the clinical implications of these signatures.
Systems biology and immunology
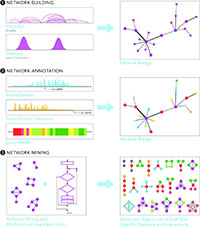 Capturing the dynamics of epigenomic patterns is essential to understand how gene expression patterns are established and maintained in healthy human cells, and how they are disrupted by pathologies. However, discovering these patterns and interpreting their biological meaning is a significant computational challenge. We tackle this challenge by developing computational tools to mine and integrate diverse data sources (ChIA-PET, ATAC-seq, RNA-seq), since intricate regulatory interactions and diverse regulatory elements cannot be inferred from a single data type. We develop machine learning models or network mining algorithms to integrate and interpret genomics data from primary human cells under the light of data accumulated in public repositories.
Capturing the dynamics of epigenomic patterns is essential to understand how gene expression patterns are established and maintained in healthy human cells, and how they are disrupted by pathologies. However, discovering these patterns and interpreting their biological meaning is a significant computational challenge. We tackle this challenge by developing computational tools to mine and integrate diverse data sources (ChIA-PET, ATAC-seq, RNA-seq), since intricate regulatory interactions and diverse regulatory elements cannot be inferred from a single data type. We develop machine learning models or network mining algorithms to integrate and interpret genomics data from primary human cells under the light of data accumulated in public repositories.
Functional genomics in Type-2-Diabetes
 In collaboration with
The Stitzel LabResearches human pancreatic islet cells and the genetic and environmental bases of type 2 diabetes.the Stitzel lab
we study genomic and epigenomic profiles in human islets and changes in these profiles associated with Type-2-Diabetes (T2D). For this we profile epigenomes/transcriptomes of human islet samples from healthy and diabetic individuals to uncover genomic signatures of T2D. We also integrate these data with genotypes of individuals via QTL analyses to uncover to what extent islet functions and responses are genetically modulated.
In collaboration with
The Stitzel LabResearches human pancreatic islet cells and the genetic and environmental bases of type 2 diabetes.the Stitzel lab
we study genomic and epigenomic profiles in human islets and changes in these profiles associated with Type-2-Diabetes (T2D). For this we profile epigenomes/transcriptomes of human islet samples from healthy and diabetic individuals to uncover genomic signatures of T2D. We also integrate these data with genotypes of individuals via QTL analyses to uncover to what extent islet functions and responses are genetically modulated.
Full Scientific Report
My laboratory’s research interests span a wide range of computational genomics activities including: i) developing low-level data processing and analysis pipelines for next-generation sequencing (NGS) datasets; ii) building data analysis, visualization and integration platforms to study epigenetic datasets in integration with other functional data (e.g., motifs; pathways; gene modules) and other NGS datasets (e.g., RNA-seq, ChIA-PET); and iii) implementing novel machine-learning applications to extract knowledge from epigenetic assays in relation to dynamic gene regulation. Specifically, I categorize my lab’s research activities into three major areas, each of which is critical for analyzing and interpreting human epigenome samples: i) putative epigenetic biomarker identification and interpretation; ii) three-dimensional chromatin interactions to identify targets of critical sites; iii) predictive epigenomic models for utilizing epigenetic patterns in human conditions.
Epigenetic biomarker identification
Genomewide association studies (GWAS) identified that a majority of the single nucleotide polymorphisms (SNPs) reside in non-coding regions of the genome. Recent epigenomic technologies such as open chromatin assays (ATAC-seq) provide an unprecedented opportunity to identify disease-causing, non-coding regulatory elements for complex human diseases. Moreover chromatin interactions assays (e.g., ChIA-PET) enable a refined interpretation of these regulatory elements. We are actively collaborating with the Stitzel, Palucka and Banchereau labs at The Jackson Laboratory for Genomic Medicine to uncover epigenetic remodeling associated with human diseases including type 2 diabetes (T2D), auto-immune diseases and cancer.
Immuno-genomics
The immune system and the repertoire of immune cells play an important role in various aspects of human health including auto-immune diseases, cancer, aging and infections. Advancements in next-generation technologies enabled genomic and epigenomic profiles from small numbers of cells, and therefore the study of immunological mechanisms under diverse conditions. We actively collaborate with the Banchereau and Palucka labs to study human immune system via generation and analysis of diverse immunogenomics data.
Three-dimensional chromatin analyses
One major challenge in interpreting the role that regulatory elements might play in human diseases is the complexity associated with the three-dimensional (3D) structure of the human genome. Genomic technologies for mapping 3D chromatin structure, i.e., ChIA-PET and HiC, have identified many regulatory elements separated by a large base-pair distance on the linear genome map that are actually in close physical proximity/contact as a result of the chromatin looping. The data generated by these technologies is the starting point from which we can begin to infer distal regulatory interactions and their system-level effects. In my lab, we build computational tools to analyze chromatin interaction datasets and integrate it with epigenetic datasets.







