Limitations to scientific progress often arise not from a lack of curiosity or imagination, but rather from the tools and technologies available. While some research questions can be sufficiently answered with a broad “hammer” approach, others require tools with greater precision and finesse. Intriguingly, a singular tool may serve as a “hammer” for some queries and as a more nuanced instrument for others.
The advent of Cre-lox recombinase technology in mouse models revolutionized the life sciences. It opened the door to elucidating gene function within a defined cell population in vivo. Cre-lox technology initially represented the pinnacle of sophistication; but as it drove scientific breakthroughs and advanced understanding, more refined questions arose, creating the need for an even more sophisticated tool.
Cre-lox technology works by defining a cell population by a single gene. However, as advancements have demonstrated, not all cells expressing a shared gene have identical roles within a biological system. This cellular heterogeneity has been challenging to investigate due to the absence of tools to selectively manipulate and examine specific cell subpopulations. Understanding the functional significance of heterogeneity within a system is essential for progressing biological knowledge, especially to identify targets for precision therapeutics and interventions.
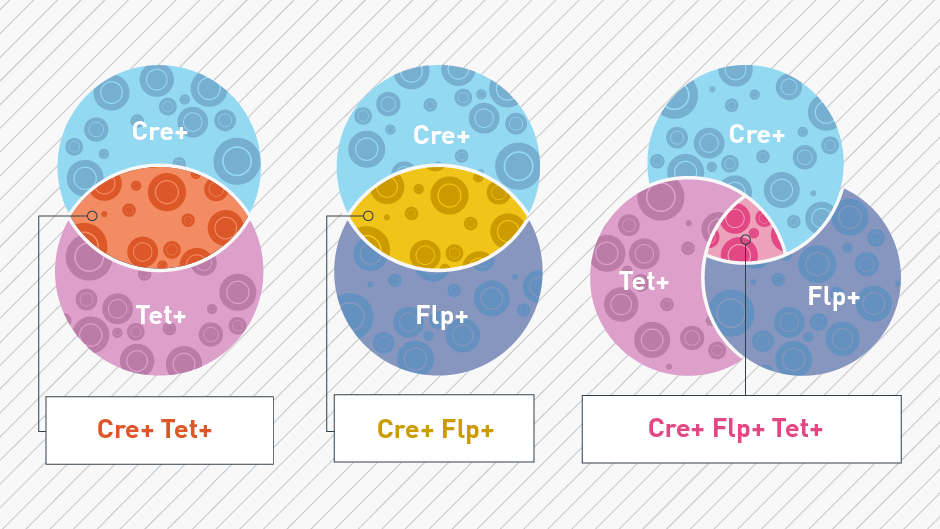
One promising approach to overcome the challenge of selectivity is through intersectional genetics. This methodology facilitates spatial and temporal genome manipulation in a more precisely defined subset of cells. Building upon the innovative Cre-lox technology, intersectional genetics combines multiple recombinase systems (Cre, CreERT, Tet, Flp, Dre) in a single mouse. Each recombinase recognizes its own target sites for recombination (Cre-lox, CreERT-lox, Tet-tTA or Tet-rtTA, Flp-Frt, Dre-rox). This specificity allows for the expression of anatomical and functional reporter proteins, gene knockouts, and gene knock-ins in cell populations defined by the expression of a distinct combination of genes, rather than a single gene.
For example, a CckCre::Ai40D or a Slc32a1Cre::Ai40D mouse enables visualization and optogenetic stimulation of all Cck-expressing cells or all GABAergic-expressing neurons, respectively. Alternatively, with an intersectional approach, a CckCre::Slc32a1FlpO::Ai80D mouse enables selective visualization and optogenetic manipulation of the specific subpopulation of interest — GABAergic neurons coexpressing Cck. The additional control of temporal manipulation can be achieved by using CreERT or Tet as one of the recombinases.
Intersectional genetics is comprised of at minimum three elements: 1) a recombinase driver line of interest, 2) a different recombinase driver line of interest, and 3) a “double stop” reporter line that is recombined by the driver lines selected. Multiple recombinase systems can be incorporated into a single mouse model through traditional breeding and/or viral delivery. Although more time-consuming, the conventional breeding approach offers the advantage of consistent, reproducible expression over successive generations. On the other hand, viral delivery provides a more expedited means of introducing multiple recombinase systems in the mouse model. This approach may be particularly advantageous when time is a critical factor, but careful consideration must be given to factors such as variability in transduction efficiency.
The JAX repository has many driver and reporter models suitable for intersectional genetics. Below is a curated list of intersectional genetics reporter models. These models can be crossed with the appropriate driver mice, enabling selective labeling, dual-labeling, and/or manipulation of well-defined cellular subpopulations.
Have questions about utilizing intersectional genetics in your research? Contact our Technical Information Services group at micetech@jax.org. Need a refresher about Cre-lox technology or recombinase systems? Click here for an excellent overview.
Cre::Tet Dual Inducible Reporter Strains
| JAX Strain # | Common Name | Promoter | Effector | Expression | Strain Nomenclature | Reference | Availability |
|---|---|---|---|---|---|---|---|
| 024108 | Ai93 | TIGRE :: TRE; CaMK2a | GCaMP6f + tTA | Forebrain calcium indicator | STOCK Igs7tm93.1(tetO-GCaMP6f)Hze Tg(Camk2a-tTA)1Mmay/J | Original | Cryo Recovery |
| 031562 | Ai162D | TIGRE :: TRE2 + CAG | GCaMP6s + tTA2s | Cre- and Tet-dependent ; calcium indicator | B6.Cg-Igs7tm162.1(tetO-GCaMP6s,CAG-tTA2)Hze/J | Original | Live |
| 030328 | Ai148D | TIGRE :: TRE2 + CAG | GCaMP6f + tTA2 | Cre- and Tet-dependent ; calcium indicator | B6.Cg-Igs7tm148.1(tetO-GCaMP6f,CAG-tTA2)Hze/J | Original | Live |
| 024115 | Ai94D | TIGRE :: TRE ; CaMK2a | GCaMP6s + tTA | Forebrain calcium indicator before dox | B6.Cg-Igs7tm94.1(tetO-GCaMP6s)Hze Tg(Camk2a-tTA)1Mmay/J | Cited (GCaMP6s) |
Cryo Recovery |
| 030217 | Ai143D | TIGRE :: TRE | RCaMP1.07 | Cre- and Tet-dependent ; calcium indicator [red] | B6.Cg-Igs7tm143.1(tetO-RCaMP1.07)Hze/J | Original | Cryo Recovery |
| 030219 | Ai139D | TIGRE :: TRE2 + CAG | EGFP + tdT + tTA2 | Cre- and Tet-dependent ; differential xFP | B6.Cg-Igs7tm139.1(tetO-EGFP,CAG-tdTomato,-tTA2)Hze/J | Original | Cryo Recovery |
| 030220 | Ai140D | TIGRE :: TRE2 + CAG | EGFP + tTA2 | Cre- and Tet-dependent ; xFP | B6.Cg-Igs7tm140.1(tetO-EGFP,CAG-tTA2)Hze/J | Original | Cryo Recovery |
| 029679 | Optopatch3 Ai155 | TIGRE :: TRE | CheRiff-eGFP :: QuasAr3-mCitrine | Cre- & Tet-dependent ; all-optical electrophysiology | B6;129S6-Igs7tm3(tetO-Optopatch3,CAG-tTA)Acoh/J | Original | Cryo Recovery |
| 031569 | Ai169D | TIGRE :: TRE2 + CAG | ASAP2s + tTA2s | Cre- and Tet-dependent ; voltage indicator | B6.Cg-Igs7tm169.1(tetO-GFP*/TPTE2*,CAG-tTA2)Hze/J | Original | Cryo Recovery |
| 034112 | Ai195 | TIGRE :: TRE2 + CAG | jGCaMP7s + tTA2 | Cre- and Tet- dependent ; EGFP calcium indicator | B6;129S6-Igs7tm195(tetO-GCaMP7s,CAG-tTA2)Tasic/J | Original | Live |
| 029633 | Rosa26-CAGs-LSL-RIK knock-in | CAG :: TRE | mKate2 + rtTA3 | Cre-inducible mKate2 fluorescence (far-red) ; then Dox-inducible (Tet-On) gene expression | B6.Cg-Gt(ROSA)26Sortm2(CAG-rtTA3,-mKate2)Slowe/J | Original | Live |
Cre::Flp Dual Reporter Strains
| JAX Strain # | Common Name | Promoter | Effector | Expression | Strain Nomenclature | Reference | Availability |
|---|---|---|---|---|---|---|---|
| 037382 | Ai224 | TIGRE :: CAG | EGFP/tdTomato | Cre-dependent EGFP expression; Flp-dependent tdTomato expression | B6.Cg-Igs7tm224(CAG-EGFP,CAG-dTomato)Tasic/J | Original | Live |
| 025109 | Ai80D | R26 :: CAG | CatCh (ChR2*L132C) / EYFP | Cre- and Flp-dependent ; opsin/xFP | B6.Cg-Gt(ROSA)26Sortm80.1(CAG-COP4*L132C/EYFP)Hze/J | Original | Cryo Recovery |
| 024846 | R26 LSL FSF ReaChR-mCitrine | R26 :: CAG | ReaChR / mCitrine | Cre- and Flp-dependent ; opsin/xFP | B6;129S-Gt(ROSA)26Sortm2.1Ksvo/J | Original | Cryo Recovery |
| 030206 | RC :: FPSit | R26 :: CAG | Synaptophysin / EGFP + tdTomato | Cre- and Flp-dependent ; xFP [synapse] | B6;129S6-Gt(ROSA)26Sortm10(CAG-Syp/EGFP*,-tdTomato)Dym/J | Original | Cryo Recovery |
| 021875 | Ai65D | R26 :: CAG | tdTomato | Cre- and Flp-dependent ; xFP | B6;129S-Gt(ROSA)26Sortm65.1(CAG-tdTomato)Hze/J | Original | Live |
| 026932 | RC :: FLTG | R26 :: CAG | tdTomato; EGFP | Flp-dependent tdT ; then Cre-dependent EGFP | B6.Cg-Gt(ROSA)26Sortm1.3(CAG-tdTomato,-EGFP)Pjen/J | Original | Live |
| 029040 | RC :: FPDi | R26 :: CAG | inhib. Gi-DREADD (hM4Di) :: mCherry | Flp-inducible mCherry, then Cre- & CNO-inducible canonical Gi pathway (neuron silencing) | B6;129S6-Gt(ROSA)26Sortm9(CAG-mCherry,-CHRM4*)Dym/J | Original | Cryo Recovery |
| 026942 | RC :: FL-hM3Dq | R26 :: CAG | eGFP :: FLEx switch hM3Dq/mCherry | Flp-inducible eGFP and RC::L-hM3Dq allele, then Cre-inducible hM3Dq-mCherry-2ACT88 fusion protein & CNO inducible hM3Dq activation (canonical Gq pathway, neuronal activity/neuronal firing) | B6.Cg-Gt(ROSA)26Sortm3.2(CAG-EGFP,-CHRM3*/mCherry/Htr2a)Pjen/J | Original | Live |
| 036590 | Cot2 / R26-EYFP | R26 :: CAG | Cot2 allele (Cre or Tomato on chromosome 2) + EGFP | Flp-dependent & tamoxifen inducible expression of Cot2, then Cre-dependent expression of EYFP (introduces mosaicism to delineate cell-autonomous functions of any gene in different tissues) | STOCK Gm10822Tn(pb-CAG-tdTomato,Cre)Cot2Zhu Gt(ROSA)26Sortm1(EYFP)Cos/J | Original | Live |
Cre::Flp Dependent, Tet Inducible Reporter Strains
| JAX Strain # | Common Name | Promoter | Effector | Expression | Strain Nomenclature | Reference | Availability |
|---|---|---|---|---|---|---|---|
| 034112 | Ai195 | TIGRE :: TRE2 + CAG | jGCaMP7s (EGFP) + tTA2 | Cre- and Flp- dependent jGCaMP7s EGFP and tTA2 expression, Dox inducible inhibition of tTA2 activity (reduce jGCaMP7s expression) | B6;129S6-Igs7tm195(tetO-GCaMP7s,CAG-tTA2)Tasic/J | Original | Live |
| 037379 | Ai211 | TIGRE :: tetO + CAG | ChrR-tdT (light-gated cation channel ; neuronal activity) +tTA2 | Cre- and Flp-dependent tTA2 and ChrR-tdT expression, Dox controllable expression of ChrR-tdT expression | B6.Cg-Igs7tm211(tetO-ChrimsonR/tdTomato,CAG-tTA2)Tasic/J | Original | Cryo Recovery |
| 037378 | Ai210 | TIGRE :: tetO + CAG | jGCaMP7f (EGFP) + tTA2 | Cre- and Flp- dependent jGCaMP7f EGFP and tTA2 expression, Dox inducible inhibition of tTA2 activity (reduce jGCaMP7f expression) | B6.Cg-Igs7tm210(tetO-GCaMP7f,CAG-tTA2)Tasic/J | Original | Cryo Recovery |