 Christine Beck (center) with her lab group (l to r): Parithi Balachandran, Denisse Tafur, Isha Walawalkar, and Alex Nesta. Photo credit: Charles Camarda
Christine Beck (center) with her lab group (l to r): Parithi Balachandran, Denisse Tafur, Isha Walawalkar, and Alex Nesta. Photo credit: Charles Camarda
When looking at genomic DNA containing billions of bases labeled A, C, G or T, it’s easy to think of them as static, linear sequences. However, genomes in living cells are anything but static.
They are active, dynamic three-dimensional structures exquisitely organized and packed into the nucleus. What is more, they constantly undergo DNA repair to fix lesions related to environmental damage and cellular processes like DNA replication.
Millions of copies of highly repetitive sequences known as transposable elements (TEs) comprise more than 50% of the human genome. They are known to contribute greatly to genetic variation and human diversity, and a small number are still able to mobilize in human genomes.
Structural variation and transposable elements
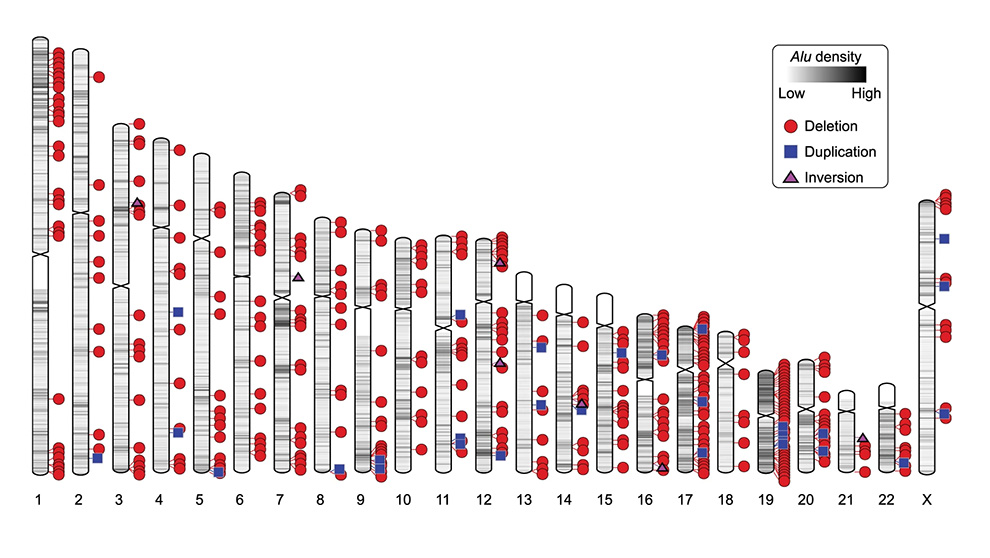
Jackson Laboratory (JAX) Assistant Professor Christine R. Beck, Ph.D., investigates how TEs can predispose human genomes to structural variants, where sections of DNA are inserted, deleted, duplicated, or inverted. Not surprisingly, the overall prevalence of repeat content in mammalian genomes means that our DNA is more subject to incorrect repair between these repeat sequences. Beck and her laboratory investigated structural variants (SVs) between TEs, examining how they are formed and the impact of these rearrangements on human genetic diversity. In “Transposable element-mediated rearrangements are prevalent in human genomes,” published in Nature Communications, Beck and her lab reveal that transposable element-mediated rearrangements (TEMRs) are responsible for ~10% of SVs within human genomes as well as providing insight into the mechanisms of their formation.
Due to well-established limitations of short-read sequencing, it is difficult or even impossible to detect all types of SVs across mammalian genomes. Advances in long-read sequencing have provided more comprehensive variant calls, but long read methods are limited by cost, sample quantity, and method development for extracting the information present in the new data types. In order to understand the capabilities and limitations of both sequencing technologies in characterizing TEMRs, Beck and her team analyzed short-read and long-read datasets from three diverse human genomes. They developed best practices for calling TEMRs in both data types, which can aid the research community in analysis of publicly available datasets, which are composed largely of short-read sequencing data. In their analyses of three genomes, they found 543 nonredundant TEMRs, including deletions, duplications, and inversions, and they were able to infer how they were formed.
The dynamics of DNA repair
A variety of DNA repair mechanisms underlie the ability of TEs to mediate structural variation. However, fully characterizing TEMR breakpoints and determining the potential mechanisms of formation is very difficult to do across a whole genome. By using multiple methods to call SVs, Beck and her team identified a total of 5,297 SVs present within non-tandem repeat regions of the genome from three diverse genomes, with an average of 3,111 per individual. Just over 10% of them, 543, had both breakpoints in homologous TEs and could be classified as TEMRs. In order to conduct mechanistic work on the TEMRs, the team focused on the two predominant classes of TEs found at breakpoints, known as Alu (397) and LINE-1 (96), for a total of 493 TEMRs. Along with 445 deletions they were able to interrogate classes of TEMRs not previously surveyed in normal human genomes, including 33 duplications, 15 inversions (4 with complexities at the junction), and 4 TEMRS that resulted in multi-copy expansions of a locus.
But how did the DNA in the TEMRs get stitched back together? The team grouped the TEMRs into two basic groups, based on the characteristics of the breakpoint junctions where the repair occurs. Most of them (390, 79.1%) were categorized as homologous repair mediated, which requires a specific DNA template sequence for relatively accurate repair. The rest (103, 20.9%) were categorized generally as derived from non-homologous events, including end-joining or replication-based mechanisms, though other DNA repair mechanisms also likely play a role in TEMR formation. Understanding the mechanisms of TEMR formation is important; more than 50% of the TEMRs were located within genic regions, though just over 5% were in coding exons. An additional 10.5% were found within five thousand base pairs of a gene. Although the effects of these genic variants remain to be seen, previous work by Beck has examined the role of Alu-mediated TEMRs in the etiology of disease.
Next steps
The annotation of 493 TEMRs in human genomes provides important insight into the mechanisms involved in forming these important structural variants and their resulting characteristics. It is clear that TEMRs play important roles in human genomes, from providing extensive variation to contributing in many ways to health and disease. The authors argue that they continue to be underestimated in human genetics, in part due to the challenges in identifying and analyzing them across mammalian genomes. Much more examination of this mechanism is needed, and expanded structural variant data sets will allow for the discovery of new biology and, most likely, novel events associated with it.